Formal description of Stochastic Reaction Networks
Stochastic Reaction Networks are Continuous-Time Markov Chains whose transitions are described via a finite set of allowed reactions, that are transformations between the items of the different species (typically of chemical nature but not necessarily so). As an example, A+B\to C is a reaction changing two individuals of type A and B into an individual of type C. The reactions can be arranged into a finite directed graph, termed reaction graph. The nodes of the reaction graph are linear combinations of species and are termed complexes. They can be regarded to as integer non-negative vectors: as an example, if the species A and B are associated with the vectors (1, 0) and (0, 1), then the complex A+B can be regarded to as (1, 1), the complex 2A can be understood as (2,0), and so on. The object of interest is the Continuous-Time Markov Chain X(\cdot) associated with the reaction graph, which keeps track of the number of items of different species over time. Whenever a reaction takes place, the state of the process changes accordingly: if the reaction y\to y' takes place at time t, then the new state is X(t) = X(t-)+y'−y, where the complexes y and y' are regarded to as vectors.
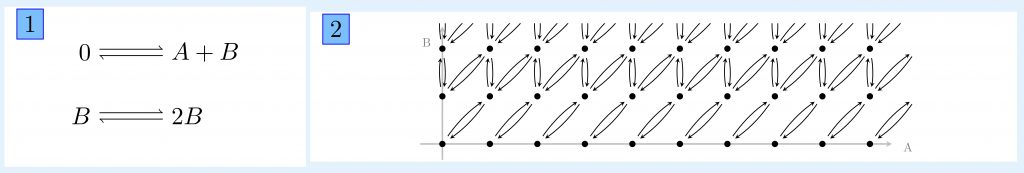
An example of reaction graph is given in panel 1. The possible transitions of the associated continuous-time Markov chain are shown in panel 2.
Often, it is assumed that the dynamics obey the mass-action law: the rate at which the reaction y\to y' takes place is proportional to the number of available combinations of molecules that can give rise to its occurrence. Namely, the rate at state x is given by
\lambda_{y\to y'}(x)=\kappa_{y\to y'}\prod_{i}\frac{x_i!}{(x_i-y_i)!}
where \kappa_{y\to y'} is a positive proportionality constant termed rate constant.
Objectives of the project
- Limit behaviour: developing new mathematical tools to study the stationary distributions of Stochastic Reaction Networks, with a particular attention to the case of mass-action models. The goal is to connect easy-to-infer properties of the reaction graph with the existence and the shape of the stationary distribution of the model, independently on the specific rate constants.
- Spatial heterogeneity: extending the classical framework introduced above by adding a spatial component to the dynamics. This component is known to play a key role in multiple biological processes. Concretely, this amounts to relaxing the homogeneity assumption on the interaction between particles, i.e. to considering particles that do not
interact with every other particle in the same way. Instead, the interactions between particles can be represented as edges of a large interaction graph. Here, the structure of the graph encodes the spatial organization and interaction between particles. The goal is to establish scaling limits for the dynamics of such interacting particle models under different
assumptions on the structure of the underlying interaction graph.